Imabalanced classification is one of the most common problems data scientist are dealing with in real life usecases. Many very common use cases such as anomaly detection, fraud detection, default prediction etc. are all imbalanced classification problems.
There are several approahces can be applied to deal with the problem: 1. Oversampling / Undersampling 2. Class Weigts 3. Custom Loss Functions
In this article I will be focusing on the third option, custom loss functions.
One of the most successful loss functions to deal with class imbalance is focal loss which was developed by Facebook AI Research in 2017. I have used this function in many imbalanced classification problems and it constantly outperformed other approaches.
The main idea is to modify cross entropy loss so that well classified majority class doesn't dominate the loss. Intuitievly, the machine learning model learns how to classify majority class instances quite well, however, due to the class imbalance the model still optimizes itself to get better at majority class (the loss function decreases more even if gets just a little better classifying majority class compared to minority class). Therefore model doesn't focus on minority class. Focal loss addresses the issue by introducing a moduling factor as can be seen below.
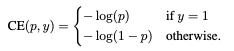
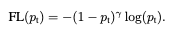
Here you might the ask "Why not just adjust the loss function according to the frequency of classes?". This approach is also commonly used, but there is a major difference between focal loss moduling factor and class weights adjustment. Class weight adjustment doesn't differentiate between easy/hard examples. With the moduling factor easyly classified examples are down weighted therefore model focuses on hard examples (mostly minority class).
In addition to moduling factor, authors also tried alpha-balanced version of the focal loss which yields slightly higher accuracy. The final version of the focal loss can be seen below.
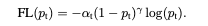
Lets try how it will work compared to other approaches. I will implement baseline model, over/under sampline model and focal loss model to compare the results.
from sklearn.datasets import make_classification
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
import lightgbm as lgb
from sklearn.metrics import classification_report
from sklearn.metrics import average_precision_score
from imblearn.combine import SMOTETomek
from scipy.misc import derivative
from sklearn.preprocessing import OneHotEncoder
Dataset
X, y = make_classification(
n_classes=2, class_sep=0.5, weights=[0.9, 0.1],
n_informative=3, n_redundant=1, flip_y=0,
n_features=20, n_clusters_per_class=1,
n_samples=5000, random_state=42
)
df = pd.DataFrame(X)
df['target'] = yy = df['target']
X = df.drop(['target'],axis=1)X_train, X_val_test, y_train, y_val_test = train_test_split(
X, y, test_size=0.3, random_state=42)
X_val, X_test, y_val, y_test = train_test_split(
X_val_test, y_val_test, test_size=0.5, random_state=42)Baseline Results with LightGBM
d_train = lgb.Dataset(X_train, label=y_train)
d_val = lgb.Dataset(X_val, label=y_val)
d_test = lgb.Dataset(X_test, label=y_test)params = {
"max_bin": 512,
"learning_rate": 0.05,
"boosting_type": "gbdt",
"objective": "binary",
"metric": "binary_logloss",
"num_leaves": 10,
"verbose": -1,
"min_data": 100,
"boost_from_average": True
}model = lgb.train(params, d_train, 1000, valid_sets=[d_val], early_stopping_rounds=50, verbose_eval=10)
Training until validation scores don't improve for 50 rounds
[10] valid_0's binary_logloss: 0.249168
[20] valid_0's binary_logloss: 0.222259
[30] valid_0's binary_logloss: 0.207472
[40] valid_0's binary_logloss: 0.200424
[50] valid_0's binary_logloss: 0.19671
[60] valid_0's binary_logloss: 0.194135
[70] valid_0's binary_logloss: 0.191952
[80] valid_0's binary_logloss: 0.189892
[90] valid_0's binary_logloss: 0.188379
[100] valid_0's binary_logloss: 0.187938
[110] valid_0's binary_logloss: 0.187842
[120] valid_0's binary_logloss: 0.187704
[130] valid_0's binary_logloss: 0.187162
[140] valid_0's binary_logloss: 0.186926
[150] valid_0's binary_logloss: 0.186899
[160] valid_0's binary_logloss: 0.186689
[170] valid_0's binary_logloss: 0.188143
[180] valid_0's binary_logloss: 0.187164
Early stopping, best iteration is:
[137] valid_0's binary_logloss: 0.186499Since we are dealing with imbalanced classifiation here, it doesn't make sense to use Accuracy or ROC-AUC. I use avereage precision score instead which is much more meaningful for imbalanced classification. I could have used f-1 or f-beta scores as well but I chose average precision since it is threshold independent.
average_precision_score(y_test, model.predict(X_test))0.592444970878653
We have 0.59 as our baseline average precision score, lets see if we can improve this
Oversampling/Undersampling
I will use SMOTETomek for Oversampling/Undersampling approach since it combines the two together.
smt = SMOTETomek(random_state=1)X_res, y_res = smt.fit_resample(X_train, y_train)d_train = lgb.Dataset(X_res, label=y_res)
d_val = lgb.Dataset(X_val, label=y_val)
d_test = lgb.Dataset(X_test, label=y_test)params = {
"max_bin": 512,
"learning_rate": 0.05,
"boosting_type": "gbdt",
"objective": "binary",
"metric": "binary_logloss",
"num_leaves": 10,
"verbose": -1,
"min_data": 100,
"boost_from_average": True
}model = lgb.train(params, d_train, 10000, valid_sets=[d_val], early_stopping_rounds=50, verbose_eval=10)
Training until validation scores don't improve for 50 rounds
[10] valid_0's binary_logloss: 0.519015
[20] valid_0's binary_logloss: 0.436783
[30] valid_0's binary_logloss: 0.393426
[40] valid_0's binary_logloss: 0.365044
[50] valid_0's binary_logloss: 0.348389
[60] valid_0's binary_logloss: 0.335367
[70] valid_0's binary_logloss: 0.32677
[80] valid_0's binary_logloss: 0.319735
[90] valid_0's binary_logloss: 0.313836
[100] valid_0's binary_logloss: 0.309641
[110] valid_0's binary_logloss: 0.306631
[120] valid_0's binary_logloss: 0.302565
[130] valid_0's binary_logloss: 0.298378
[140] valid_0's binary_logloss: 0.294932
[150] valid_0's binary_logloss: 0.293326
[160] valid_0's binary_logloss: 0.29132
[170] valid_0's binary_logloss: 0.288407
[180] valid_0's binary_logloss: 0.286276
[190] valid_0's binary_logloss: 0.283667
[200] valid_0's binary_logloss: 0.281751
[210] valid_0's binary_logloss: 0.280354
[220] valid_0's binary_logloss: 0.280005
[230] valid_0's binary_logloss: 0.278381
[240] valid_0's binary_logloss: 0.277043
[250] valid_0's binary_logloss: 0.276011
[260] valid_0's binary_logloss: 0.274862
[270] valid_0's binary_logloss: 0.273153
[280] valid_0's binary_logloss: 0.270512
[290] valid_0's binary_logloss: 0.269502
[300] valid_0's binary_logloss: 0.270204
[310] valid_0's binary_logloss: 0.269744
[320] valid_0's binary_logloss: 0.269011
[330] valid_0's binary_logloss: 0.269219
[340] valid_0's binary_logloss: 0.267952
[350] valid_0's binary_logloss: 0.267307
[360] valid_0's binary_logloss: 0.266693
[370] valid_0's binary_logloss: 0.265973
[380] valid_0's binary_logloss: 0.26577
[390] valid_0's binary_logloss: 0.265747
[400] valid_0's binary_logloss: 0.264776
[410] valid_0's binary_logloss: 0.263674
[420] valid_0's binary_logloss: 0.263634
[430] valid_0's binary_logloss: 0.263462
[440] valid_0's binary_logloss: 0.263334
[450] valid_0's binary_logloss: 0.263705
[460] valid_0's binary_logloss: 0.26252
[470] valid_0's binary_logloss: 0.26227
[480] valid_0's binary_logloss: 0.262205
[490] valid_0's binary_logloss: 0.260808
[500] valid_0's binary_logloss: 0.260792
[510] valid_0's binary_logloss: 0.260445
[520] valid_0's binary_logloss: 0.259891
[530] valid_0's binary_logloss: 0.259878
[540] valid_0's binary_logloss: 0.259983
[550] valid_0's binary_logloss: 0.258996
[560] valid_0's binary_logloss: 0.258527
[570] valid_0's binary_logloss: 0.258384
[580] valid_0's binary_logloss: 0.257741
[590] valid_0's binary_logloss: 0.257963
[600] valid_0's binary_logloss: 0.256561
[610] valid_0's binary_logloss: 0.256218
[620] valid_0's binary_logloss: 0.255757
[630] valid_0's binary_logloss: 0.255567
[640] valid_0's binary_logloss: 0.256395
[650] valid_0's binary_logloss: 0.257255
[660] valid_0's binary_logloss: 0.257816
[670] valid_0's binary_logloss: 0.257454
Early stopping, best iteration is:
[629] valid_0's binary_logloss: 0.255566average_precision_score(y_test, model.predict(X_test))0.6063398773458056
With SMOTETomek we improved average precision score from 0.592 to 0.606
Focal-loss
d_train = lgb.Dataset(X_train, label=y_train)
d_val = lgb.Dataset(X_val, label=y_val)
d_test = lgb.Dataset(X_test, label=y_test)from scipy.misc import derivative
def focal_loss_lgb(y_pred, dtrain, alpha, gamma, num_class):
"""
Focal Loss for lightgbm
Parameters:
-----------
y_pred: numpy.ndarray
array with the predictions
dtrain: lightgbm.Dataset
alpha, gamma: float
See original paper https://arxiv.org/pdf/1708.02002.pdf
num_class: int
number of classes
"""
a,g = alpha, gamma
y_true = dtrain.label
# N observations x num_class arrays
y_true = np.eye(num_class)[y_true.astype('int')]
y_pred = y_pred.reshape(-1,num_class, order='F')
# alpha and gamma multiplicative factors with BCEWithLogitsLoss
def fl(x,t):
p = 1/(1+np.exp(-x))
return -( a*t + (1-a)*(1-t) ) * (( 1 - ( t*p + (1-t)*(1-p)) )**g) * ( t*np.log(p)+(1-t)*np.log(1-p) )
partial_fl = lambda x: fl(x, y_true)
grad = derivative(partial_fl, y_pred, n=1, dx=1e-6)
hess = derivative(partial_fl, y_pred, n=2, dx=1e-6)
# flatten in column-major (Fortran-style) order
return grad.flatten('F'), hess.flatten('F')
def focal_loss_lgb_eval_error(y_pred, dtrain, alpha, gamma, num_class):
"""
Focal Loss for lightgbm
Parameters:
-----------
y_pred: numpy.ndarray
array with the predictions
dtrain: lightgbm.Dataset
alpha, gamma: float
See original paper https://arxiv.org/pdf/1708.02002.pdf
num_class: int
number of classes
"""
a,g = alpha, gamma
y_true = dtrain.label
y_true = np.eye(num_class)[y_true.astype('int')]
y_pred = y_pred.reshape(-1, num_class, order='F')
p = 1/(1+np.exp(-y_pred))
loss = -( a*y_true + (1-a)*(1-y_true) ) * (( 1 - ( y_true*p + (1-y_true)*(1-p)) )**g) * ( y_true*np.log(p)+(1-y_true)*np.log(1-p) )
# a variant can be np.sum(loss)/num_class
return 'focal_loss', np.mean(loss), Falsefocal_loss = lambda x,y: focal_loss_lgb(x, y, 0.25, 2., 2)
eval_error = lambda x,y: focal_loss_lgb_eval_error(x, y, 0.25, 2., 2)
params = {"num_class":2,
"max_bin": 512,
"metric": None,
"objective": None,
"learning_rate": 0.05,
"boosting_type": "gbdt",
"num_leaves": 10,
"verbose": -1,
"min_data": 100,
"boost_from_average": True
}# model = lgb.train(params, lgbtrain, fobj=focal_loss)
model = lgb.train(params, d_train, valid_sets=[d_val], early_stopping_rounds=20, fobj=focal_loss, feval=eval_error, verbose_eval=10,
)Training until validation scores don't improve for 20 rounds
[10] valid_0's focal_loss: 0.0552852
[20] valid_0's focal_loss: 0.0396717
[30] valid_0's focal_loss: 0.0314231
[40] valid_0's focal_loss: 0.0270093
[50] valid_0's focal_loss: 0.0245737
[60] valid_0's focal_loss: 0.0231538
[70] valid_0's focal_loss: 0.0223817
[80] valid_0's focal_loss: 0.0219951
[90] valid_0's focal_loss: 0.0217383
[100] valid_0's focal_loss: 0.0215834
Did not meet early stopping. Best iteration is:
[100] valid_0's focal_loss: 0.0215834In this implementation I have used alpha=0.25 and gamma=2.0 which are the default choices made by the authors and they are both good defaults.
pred_df = pd.DataFrame(model.predict(X_test))
pred_df = pd.DataFrame(np.exp(pred_df))
pred_df['sum'] = np.sum(pred_df,axis=1)
pred_df[0] = pred_df[0]/pred_df['sum']
pred_df[1] = pred_df[1]/pred_df['sum']
pred_df = pred_df.drop('sum',axis=1)
probas = np.array(pred_df[1])average_precision_score(y_test, probas)0.6338175201422622
With focal-loss we improved the average precision score to 0.633 which is significantly higher than two other scores we have got so far. If you would like to learn more about focal-loss please have a look at the original paper here